Data ingestion
Data ingestion consists of 3 steps:
- Transforming metadata (including phenotypic data).
- Transforming genomic variations (VCF).
- Load data into MongoDB.
Important
Here we will give you an overview of the tree steps. For an in depth explanation, please follow this tutorial.
1 - Transforming metadata (including phenotypic data)
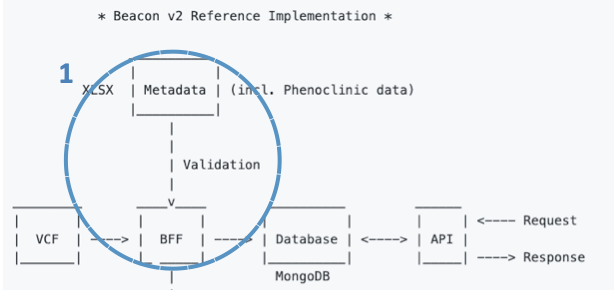
The overall idea is that the B2RI will facilitate transforming your data (sequencing methodology, bioinformatics tools, phenotypic data, etc.) to the format of the Beacon v2 Models.
The Beacon v2 Models define the default data structure (or schema) for the biological data responses. The Models are defined using JSON Schema and the information is structured in a hierarchical form. The schemas consist of multiple properties or terms (a.k.a., objects).
We have chosen MongoDB as a de facto database as it works directly with JSON files. This way, we can store the data directly in the database according to the Beacon v2 Models and provide responses (Beacon v2 compliant) without the need of re-mapping the data at the API level.
About alternative response schemas for biological data
A priori, Beacon v2 specification allows for alternative schemas for the responses (e.g., Phenopackets). At this time (Apr-2022), this option is not supported by the Beacon v2 API.
2 - Transforming genomic variations (VCF)
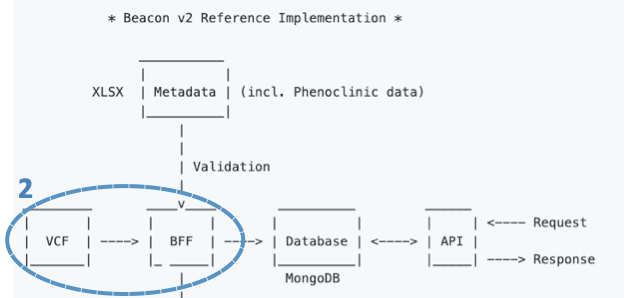
For genomic data, the B2RI has a tool that takes as input a VCF file and uses BCFtools and SnpEff and SnpSift to annotate it. Once annotated, the tool transforms VCF data to the genomicVariations entry type in the Beacon v2 Models and serializes it to a JSON file.
3 - Load data into MongoDB
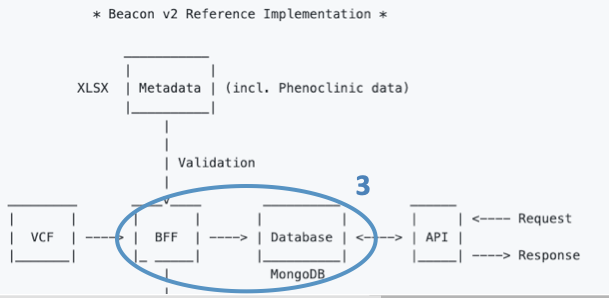
Once transformed, the JSON files conform what we call the Beacon Friendly Format (BFF).
The last step is loading the BFF files into a MongoDB instance.
The B2RI has a tool that loads BFF files to MongoDB. Once loaded, we will refer to them as collections.
Included utilities
The data ingestion tools include a few utilities that will help you with data processing and beyond:
BFF Genomic Variations Browser
Important
BFF Genomic Variations Browser is not a full UI for Beacon v2 as it does not allow for cross-queries to other collections (e.g., individuals).
BFF Genomic Variations Browser enables user-friendly visualization of genomicVariations documents (stored as a JSON array) via dynamic tables embedded in HTML.
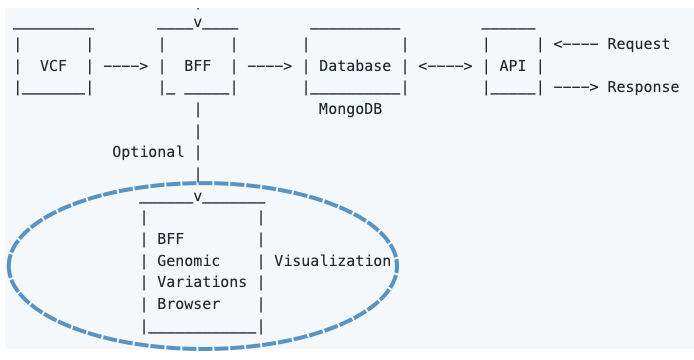
The browser's developer has first-hand experience in Clinical Genomics and thought that the HTML will be a nice addition for some users.
BFF Genomic Variations Browser only displays a subset o variants (i.e., those having HIGH value on the Annotation Impact field).
The variants are displayed as HMTL-tabs from gene lists (a.k.a. gene panels). These gene panels are plain text files consisting of one column (name of genes). The extension is '.lst'. By default, they're located under $beacon_path/browser/data, but you can specify a different location via the parameter paneldir in the config.yaml file. Feel free to create and add your own panels.
BFF Genomic Variations Browser will display the filtered variants according to all .lst files in paneldir folder.
The resulting HTML are local. They load a local JSON file and display it as a searchable table.
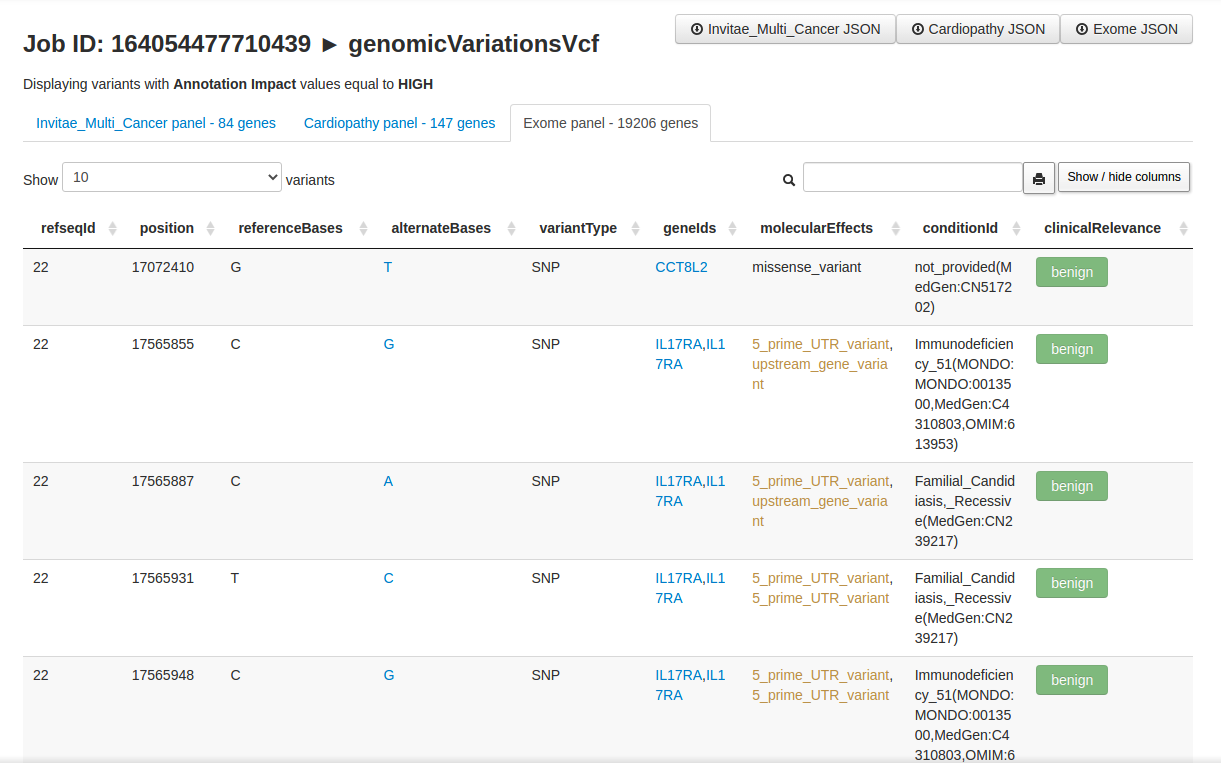
The table allows for columns re-ordering and the search box accepts complex regex e.g. rs12(3|4) (tp53|ace2) splice.
Now, importing local JSON files (w/ AJAX) is restricted by web browsers (by default). It's a security thing and makes sense most of the times. However, it has no sense here and thus we will bypass this "functionality". It's harmless.
To overcome this issue we provide several alternatives, ranked by the level of difficulty.
1 - The simplest option is to use chromium-browser from the command line and add the flag --allow-file-access-from-files, like this:
$ sudo apt install chromium-browser # To install chromium in Debian-based distribution
$ cd beacon_XXX/browser # where XXX is the job ID
$ chromium-browser --allow-file-access-from-files XXX.html
2 - The second simplest option is to use firefox and disable the restriction with the boolean privacy.file_unique_origin located in about:config (address bar).
3 - The third option is to load the HTML via http(s) protocol. There are quite a few ways of doing this (w/o resorting to apache2/nginx).
Below I am displaying a few:
- With Php
$ php -S localhost:8000
- With Ruby
$ ruby -run -e httpd . -p 8080
- With Python 3
python3 -m http.server
- Others
Mojolicius, Node.js and other web frameworks also allow for this.